
- UC San Diego team trained AI to construct a new model of the cell.
- Model detects new protein communities and predicts their functions.
- The technique revealed dozens of novel cell components.
A new study by UC San Diego researchers combined machine learning with protein imaging and biophysical association to create a map of subcellular components. The AI generated map, called Multi-Scale Integrated Cell (MuSIC), revealed 69 subcellular systems—around half of which are new, undocumented cell components. The technique, detailed in a November 24, 2021, article in Nature [1], resulted in a map that looks nothing like the cell diagrams in biology textbooks.
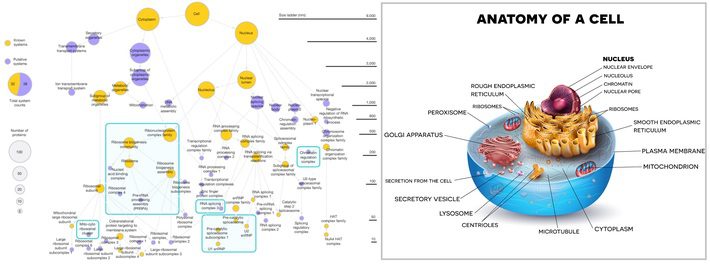
In the MuSIC image on the left, known cell components are in gold, new cell components are in purple, and arrows indicate containment of the lower system by the upper system. The system doesn’t map the cell component to a specific place, like those in the classic diagram on the right, partly because their locations are fluid, changing with cell type and situation [2]. In other words, the cell isn’t made up of neatly placed components suspended in intracellular fluid; there is a hierarchy of biological order, where a nested succession of processes determines the functional and spatial organization of cells [3].
Traditional imaging research tends to focus on physical size and distances between cellular components, but this new AI-based research suggests that protein interactions can give a complementary measure of intracellular distance. [1] If you think of intercellular organization as being like a small city, a whole host of inputs make up a dynamic object that’s much more than buildings and streets. For example, people’s movements are a function of factors like rush hour traffic, social interactions, or weather. In the same way, the full features of cells can’t be accurately described by a two-dimensional map; they are governed by a myriad of complex biological processes that occur within the cell walls.
Combining Traditional Techniques and AI
Finding a bridge to span the gap from nanometer to micron scale had—up until this recent study—eluded researchers in the biological sciences. “Turns out you can do it with artificial intelligence,” says Trey Ideker, PhD, one of the study leaders, in a UCSD press release [2].
Cellular components are usually mapped with either biophysical association or microscope imaging. Both techniques have their limitations: super-resolution microscopes, which can see inside cells with resolution better than 250 nanometers [4] are limited by the wavelength of the electron beam [5]; biochemistry techniques can map structures further down the nanometer scale, but still can’t see cell structures on the micron level (one micron is 1/1000th of a nanometer). The two approaches generate massive amounts of data with distinct qualities and resolutions that are usually analyzed separately [6]. With machine learning, the ability to analyze both sets of data comes into play. This new technique combines the traditional imaging methods with deep learning to map cell data from multiple sources, including cellular microscopy images.
The Procedure
The study began with a matched dataset of immunofluorescence cell images, including human embryonic kidney cells with 661 proteins. Deep neural networks embedded each protein and assigned them coordinates in reduced dimensions. Distances between the proteins was calculated and calibrated with a reference set of known cellular components of known or estimated diameter. A supervised ML model (random forest regression) was trained to estimate the distance of any protein pair from its embedded coordinates. After all the distances were analyzed, the MuSIC 1.0 hierarchy was created, with 69 protein communities—54% of which had never been categorized before.
The Future of Cell Maps
Ideker noted this pilot study looked at just 661 proteins from one cell type. The map is being developed to cover all human proteins, a huge task that may result in a unified map of cellular components or in separate maps for different cell types. Identification of new protein communities brings more promise to the hope of curing cancer and other diseases that start at the intercellular level. “Eventually we might be able to better understand the molecular basis of many diseases,” Ideker said, “…by comparing what’s different between healthy and diseased cells” [2].
References
Cell image: Adobe Creative Cloud [Licensed]
[1] A multi-scale map of cell structure fusing protein images and interactions
https://www.nature.com/articles/s41586-021-04115-9.epdf
[2] https://health.ucsd.edu/news/releases/Pages/2021-11-24-we-might-not…
[3] https://journals.asm.org/doi/full/10.1128/MMBR.69.4.544-564.2005
[4] https://news.mit.edu/2015/enlarged-brain-samples-easier-to-image-0115
[5] https://learn.genetics.utah.edu/content/cells/scale/
[6] https://www.biorxiv.org/content/10.1101/2020.06.21.163709v1?
